High-specificity proximity ligation assay to enable detection with low-affinity agents
Technology description
The invention is a method that enables compatibility with low quality capture agents (e.g. antibodies displaying high equilibrium dissociation constant, KD) in proximity-based detection methods and improves assay performance for high quality capture agents.
Researchers at Stanford’s Genome Technology Center have developed a highly sensitive, straightforward, automatable proximity ligation assay (PLA) method to reduce background and improve quantitative detection of protein biomarkers. Up until now, a major drawback of PLA has been that it does not work with low affinity antibodies due to high levels of background noise. This invention, circular PLA (c-PLA), greatly reduces the background by using two oligonucleotide probes that ligate to form circular DNA. This both lowers the probability of random background from the ligation reaction and enables exonuclease clean-up of residual uncircularized DNA. Together, this increases the sensitivity and specificity of the assay to improve overall performance (limit-of-detection, dynamic range) which thereby enables the use of low-affinity capture reagents. Furthermore, the straightforward workflow is performed in a single reaction tube with a small sample volume making it amenable to automation, multiplexing and high throughput assay development. This technology could expand the utility of PLA for a variety of research and diagnostic applications where good affinity reagents are not available.
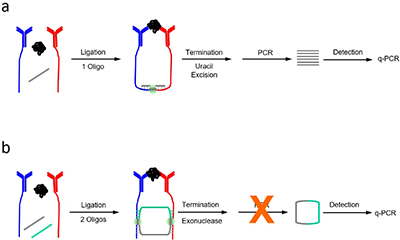 Traditional vs. circular PLA.a) Traditional proximity ligation assay (t-PLA) detects proteins using pairs of antibody-DNA conjugates (red and blue) brought into close proximity when they bind a target analyte. Detection is aided by a single oligonucleotide creating a ligation product that is preamplified before quantifying with qPCR. b) In circular proximity ligation assay (c-PLA), the antibody-tethered oligonucleotides act as bridges for two ligation events which form a circular ligation product. This lowers the probability of random background ligation events resulting in increased specificity. Exonuclease treatment further reduces background by degrading all uncircularized DNA thereby eliminating the preamplification step before quantification.
Traditional vs. circular PLA.a) Traditional proximity ligation assay (t-PLA) detects proteins using pairs of antibody-DNA conjugates (red and blue) brought into close proximity when they bind a target analyte. Detection is aided by a single oligonucleotide creating a ligation product that is preamplified before quantifying with qPCR. b) In circular proximity ligation assay (c-PLA), the antibody-tethered oligonucleotides act as bridges for two ligation events which form a circular ligation product. This lowers the probability of random background ligation events resulting in increased specificity. Exonuclease treatment further reduces background by degrading all uncircularized DNA thereby eliminating the preamplification step before quantification.
Additional Information
http://med.stanford.edu/sgtc/
Application area
Protein detection- PLA-based assay to detect low quantities of analytes (e.g., proteins, peptides, drugs, metabolites) in blood with end user applications in research and diagnostics
Advantages
High specificity with low background- increases capture-probe concentration and signal-to-noise ratio compared conventional PLA techniques which thereby:enablesdetection using low affinity capture agents(e.g., antibodies, or aptamers with high equilibrium dissociation constant) lowers limit of detection and widens dynamic range (down to femtomolar concentration and up to five orders of magnitude dynamic range) decreases assay variability
Straightforward workflowcompatible withmultiplexing and high throughput/automatedapproaches:performed in single reaction tube with small (2 uL) sample volume eliminates preamplification step prior to quantification no pre-concentration on solid phase variety ofoptions for quantification(e.g., digital, fluorescent, electrical and more)
Kinetic analysis- modeling assay conditions for performance provides a tool to screen for suitable capture agents and optimize detection

